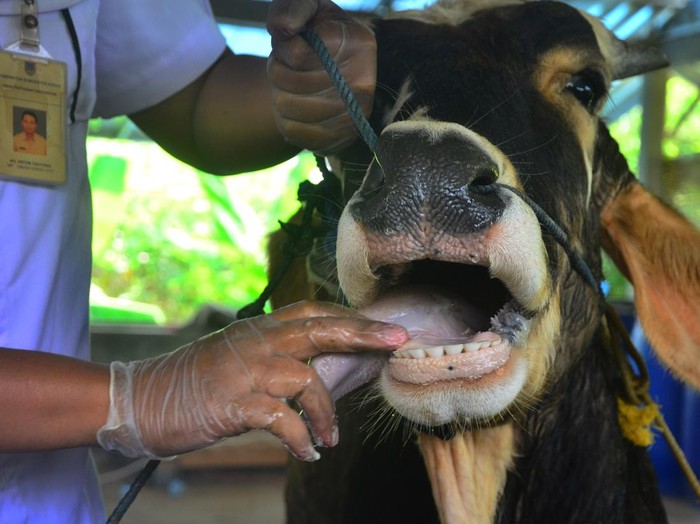
Foot and Mouth Disease Virus (FMDV) is one of the important diseases in animals, especially cloven-hoofed animal with a high transmission rate. According to OIE, FMDV is included in the list of infectious animal diseases with a 100% morbidity rate in animal populations with confirmed by FMDV and mortality in young animals reaching 20-30% while in adult animals reaching 1-5% %. FMDV outbreaks are common in Southeast Asia, including in Indonesia. First FMDV outbreak was found in Indonesia on April 28th 2022, with a total of 1,247 cases in East Java (Gresik, Lamongan, Sidoarjo, Mojokerto) and 1,881 cases in Aceh Tamiang District, Aceh Province. This was stated in the Decree of the Minister of Agriculture of the Republic of Indonesia concerning the determination of FMDV outbreak areas in several Regencies in East Java and Aceh Tamiang District, Aceh Province. FMDV outbreaks in Indonesia were included in cases of re-emerging disease, where in 1884 Indonesia had experienced FMDV cases and in 1990 Indonesia was declared FMDV free.
FMDV is caused by a virus from the genus Apthovirus in the Picornaviridae family and have seven FMD serotypes (O, A, C, SAT-1, SAT-2, SAT-3, and Asia1) in which the FMD-causing virus has a high degree of genetic and antigen diversity. The 25-30 nm virus particle has an icosahedral capsid composed of proteins, non-envelope with a genome of positive-sense single-stranded RNA encoding 4 structural proteins and non-structural proteins. Structural proteins are composed of VP1-VP4 while non-structural proteins consist of 28. 2C, 3A 3B, 3C and 3D. VP1 GH Loop protein, which is not shared by other proteins, contains RGD (arginine-glycine-aspartic acid) peptide sequences that play a role in receptor binding. Besides serving as a determinant of virus serotype classification, the VP1 gene is also used to design primers for amplification and sequencing protocols for FMD virus detection.
In 2001 the O/ME-SA/Ind-2001 lineage was first reported in India and thereafter spread across several countries such as North Africa, Middle East Asia, Southeast Asia, and East Asia. The O/ME-SA/Ind-2001 lineage has been classified into sub-lineages a, b, c, d, and e, with O/ME-SA/Ind-2001e being found mostly in Southeast Asia, including Indonesia. FMDV O/ME-SA/Ind-2001e were circulated in Indonesian and the most common causative agent spread in Cambodia, Myanmar, Bangladesh and Thailand.
This study aims to detect FMD based on clinical symptoms from field cases in beef cattle using reverse transcriptase-polymerase chain reaction (RT-PCR) method followed by sequencing. The results of this study are expected to provide information and can be used in policy and control of FMD virus to overcome losses due to FMD disease.
Materials and Methods
Samples were collected from cattle diagnosed with FMD with clinical symptoms of fever and vesicles in the mouth and feet in August 2022. Samples were collected from vesicle swabs on lesions in the mouth and feet of beef cattle from smallholder farms in Banyuwangi, East Java, Indonesia. Samples were collected with Phosphate Buffer Saline (PBS) with 5% antibiotic transport medium. Samples were stored at -80°C in the laboratory before analysis.
The extraction of viral RNA from the specimens using QIAmp Viral RNA mini kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). RNA was eluted with 60 µl of RNase- and DNase-free water and stored at -80°C before further analysis.
RT-PCR has performed for detecting and genotyping of FMDV using the one- step RT-PCR PCR (AMV) Takara Kit as previously described. First detection of FMDV infection using universal primers and to determine serotype O in the VP1 gene using one pair of primers are showen in Table 1. The primers were synthesized by (Macrogen Co., Seoul, South Korea). Total 2.5 µl RNA sample eith 8 µl nucleas-free water, 2.5 µl, forward primer (4pmol/ µl), 5 µl reverse primer (4pmol/ µl), 4.2 µl, MgCl2 2.5 µl, dNTP Mix 1. 25 µl, 10x one step buffer 1.25 µl, AMV r-tase 0.25 µl, AMV optimized Taq 0.25 µl, and RNAse inhibitor 0.25 µl were put into a PCR tube with the final mixture was 15 µl and then inserted into a conventional PCR machine. One-step RT-PCR cycle conditions were 30 minutes at 42⁰C followed by 5 minutes of 94⁰C inactivation, then at cycle 35 with conditions of denaturation 94⁰C for 60 seconds, anelling 55.5⁰C for 60 seconds, elongation 72⁰C for 60 seconds, and final extension 72⁰C for 10 minutes. RT-PCR products were stored in a refrigerator before electrophoresis.
Results and Discussion
RT-PCR tests on suspect samples were conducted to confirm FMD virus infection causing productivity decline in the livestock sector. The universal primer set was used for first-time detection in cattle suspected of FMDV infection. RT-PCR product amplification result according to the expected size (328bp) is considered as a positive result. The results of this study are consistent with previous studies that RT-PCR was used to detect FMDV for primary diagnosis with a set of universal 1F/1R primers with an expected band of 328bp. A pair of IF/IR-based RT-PCR universal primers can be used in rapid detection of FMD infected animals. Same samples were detected using serotype O specific primers on the VP1 gene by RT-PCR. The result showed the presence of DNA bands in the three samples. The VP1 gene fragment was obtained with visualization of DNA bands with a length of 1165 bp.
RT-PCR is a highly sensitive and specific method that has been used to detect FMDV outbreaks or re-infections disease. The difference between two RT-PCR results with the same sample suggests that the universal primer set more sensitive to detect FMD viral infection. Universal primers encoding the untranslated region (UTR) more sensitive in detecting O, A and Asian 1 serotype viruses. The FMDV viral genome contains one open reading frame (ORF) that begins and ends with 5′ and 3′ untranslated regions (UTR). The genome of FMDV contains several functional elements including in the untranslated region (UTR), in the 5′ UTR the internal ribosome entry site (IRES) element initiates cap-independent translation while the 3′ UTR is for stimulating IRES activity.
The VP1 belonged to structural protein that serves as capsid protein on FMDV and primary antigen that elicits and neutralizes antibody responses. VP1 gene as a determinant of virus serotype classification, serotype-specific primer design and as a determinant of conserve regions in the GH Loop due to the presence of the amino acid RGD (arginine-glycine-aspartic acid). RT-PCR results show positivity in the VP1 gene less than the UTR, this can be caused by several factors. The primer design used to detect FMDV serotype O in the VP1 gene is less sensitive, it is necessary to design own primers from local isolates that have high accuracy. Nucleotide and amino acid changes in the VP1 serotype O gene also affect the primers, the presence of mutations, substitutions, deletions and insertions causes sensitive primers. Focus on sequencing the whole viral genome to reveal possible mutations at the GH Loop site of the serotype O-specific primers of the VP1 gene, and then designing primers to target this site.
In 1889, Indonesia experienced an FMD outbreak that spread throughout the region, and was declared FMD-free in 1990. For 32 years Indonesia was free from FMD until finally in 2022, FMD outbreaks were again found in several regions in Indonesia. Meat imports and illegal entry of animals are suspected to be one of the sources of FMD virus spread in Indonesia. Foot and mouth disease is of great importance to livestock breeding due to the rapid spread of the virus. It should be recognized and diagnosed as soon as possible for outbreak control and management. Rapid and accurate detection of FMDV is necessary for effective outbreak control in planning and country preparedness in the face of outbreaks.
In this study, we investigated cattle suspected of FMDV infection using one step RT-PCR method. These findings indicate the importance of surveillance studies and genetic analysis to monitor FMD virus to evaluate the emergence of genetic variation in Indonesia. Future studies related to genetic analysis of FMD viruses from field isolates in Indonesia are needed. Monitoring of FMD outbreaks and the spread of O/ME-SA/Ind-2001e in Indonesia should be conducted to evaluate the national spread and establish efficient control strategies.
Conclusion
Based on the results of this study, it can be concluded that the RT-PCR technique can accurately detect FMD virus infection in cattle on smallholder farms in Banyuwangi, East Java, Indonesia. This study found that universal primers in the UTR region showed more sensitive results compared to serotype O specific primers of VP1 gene. The RT-PCR protocol can be used to handle samples in a single assay and can be a valuable tool to complement routine diagnosis of FMD viruses from field outbreaks and prevention strategies.
Authors: Zayyin Dinana